A bitter aftertaste: How nitrogen gets into the bitter substances of Solanum plants
An enzyme originally derived from basic plant metabolism controls the synthesis of steroidal glycoalkaloids, important defense substances in tomatoes.
A team of researchers from the Max Planck Institute for Chemical Ecology has succeeded in characterizing the decisive biosynthetic step in the production of steroidal glycoalkaloids, important defense substances in various Solanum plants.

Solanum plants such as tomatoes, potatoes and eggplants produce nitrogen-containing defense substances, known as steroidal glycoalkaloids, which protect them from herbivores and plant diseases. The incorporation of nitrogen is a key factor in the toxicity and thus bioactivity of steroidal glycoalkaloids.
Previous studies have already identified several enzymes involved in the biosynthesis of steroidal glycoalkaloids in tomato and potato. Although the GLYCOALKALOID METABOLISM12 (GAME12) enzyme investigated in the current study was earlier shown to be involved in steroidal glycoalkaloid biosynthesis, the predicted biochemical reaction that incorporates nitrogen atom into the steroidal backbones leading towards production of the defensive compounds remained unknown. Scientists from the Department of Natural Product Biosynthesis at the Max Planck Institute for Chemical Ecology in Jena have now revealed how GAME12, a transaminase enzyme, hijacked from its role in basic plant metabolism eventually directs the production of steroidal glycoalkaloids in Solanum plants.

"Characterization of this central reaction in steroidal glycoalkaloid production was challenging as the proposed substrate of the reaction was never confirmed experimentally”, says Dagny Grzech, the first author of the publication. “In the study, we used a combination of biochemistry and metabolomics to demonstrate the activity of GAME12- the key enzyme of this pathway”.
In cooperation with scientists from the Lichman group based at the University of York, UK and the Aharoni group from the Weizmann Institute of Science, Israel researchers showed that the crucial enzyme evolved from an ancestor involved in basic plant metabolism. “It has been known that GAME12 belongs to the γ-aminobutyric acid transaminase (GABA-T) enzyme family. GABA-T proteins play a crucial role in the basic plant metabolism in mitochondria, where they catalyze an important reaction - a bypass of the central carbon cycle that forms the basis for all plant life functions. Our study shows that GAME12 evolved from an ancestral GABA-T enzyme through neofunctionalization- a process in which a gene acquires a novel function following a gene duplication event. Notably, this hijacking of core GABA enzyme was achieved via the change of the localization of the protein into a different cell compartment and changes in the overall architecture of the protein”.
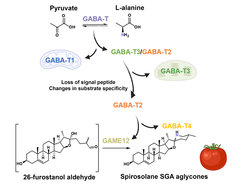
“Genomics analysis coupled with extensive biochemical characterization allowed us to hypothesize how GAME12 evolved from its core metabolism ancestor. This grants significant insight into how enzymes involved in plant specialized metabolism come into their specific roles”, says Prashant Sonawane from the Department of Natural Product Biosynthesis at the Max Planck Institute for Chemical Ecology (recently moved to the Department of Biochemistry, University of Missouri, Columbia, MO, USA), who conceived and supervised the study. “Our results also provide basis for engineering of steroidal glycoalkaloid production in plant systems. We demonstrated that transferring this GAME12 enzyme alone into the leaves of the black nightshade plants (Solanum nigrum) is enough for the production of nitrogen-containing steroidal glycoalkaloids, which are otherwise completely absent in the leaves. This re-direction of metabolic flux could potentially be achieved in other agriculturally relevant plants, which produce non-nitrogenous, steroidal defensive compounds such as saponins”.
Sarah O'Connor, head of the Department of Natural Product Biosynthesis, sees the results in a broader context: "Our study shows that biosynthetic pathways display great plasticity, which can be manifested not only by the changes in the catalytic activities of the involved enzymes but also by the alterations to their spatial organization. Such ‘metabolic’ plasticity results in the enormous diversity of plant specialized metabolic pathways."


